近日,Nucleic Acids Research杂志在线发表了华中农业大学生命科学技术学院周志鹏教授课题组、华中科技大学王锐教授课题组和纽约州立大学奥尔巴尼分校Jia Sheng教授课题组的共同研究成果,论文题目为“Bioorthogonal Labeling and Profiling of N6-isopentenyladenosine (i6A) Modified RNA”。该论文首次报道在单碱基分辨率定量检测i6A修饰的方法,为揭示i6A修饰的生物学功能和分子机制提供了强大的工具。

tRNA反密码子环第37位腺苷酸的N6-isopentenyladenosine (i6A37)是最早发现的tRNA修饰之一,存在于细菌和真核生物中部分用于解码UNN密码子的tRNA上。在真核生物中,i6A37由Trit1或其同源蛋白催化形成。该修饰通过影响翻译效率和翻译保真度参与众多生物学过程的调控。在人中,TRIT1基因是必须基因,其突变可能引起线粒体疾病以及癌症等疾病发生。然而,过去几十年对i6A37修饰的研究进展较为缓慢,原因之一在于缺乏高效精确的检测方法。
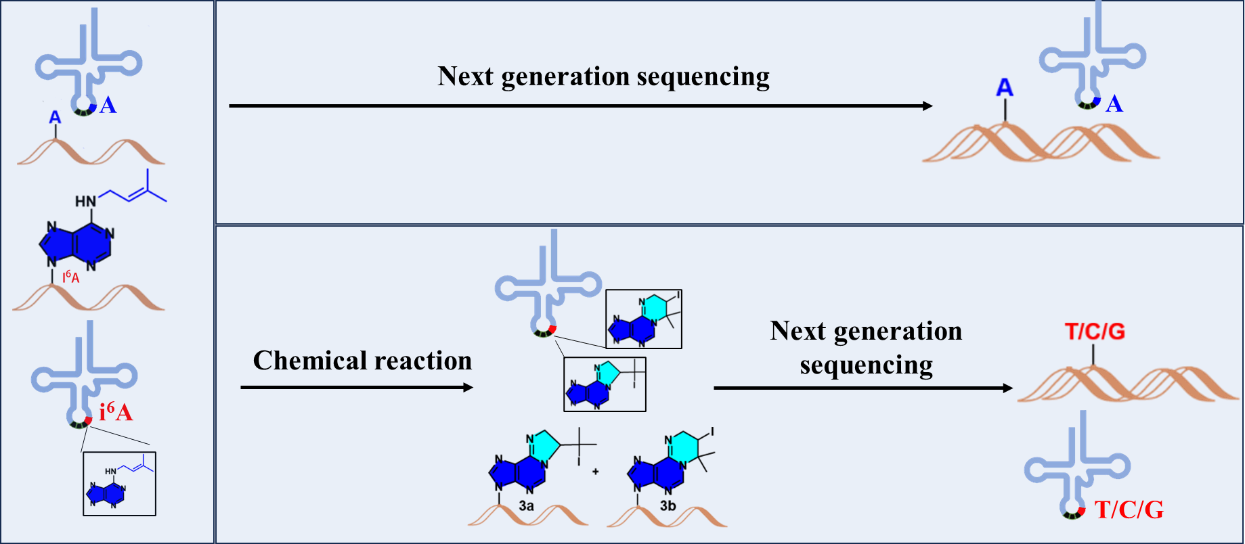
在本研究中,科研人员利用碘单质处理RNA,使i6A的烯丙基环化将其由氢键的受体转化为氢键的供体,从而导致反转录时发生错配。经过高通量测序,科研人员可以根据错配的位置确定i6A的修饰位点、根据错配发生的频率确定i6A的修饰水平。科研人员将此方法命名为IMCRT tRNA-seq,并成功利用该方法检测了芽殖酵母的i6A37修饰状况,发现6个细胞质tRNA和3个线粒体tRNA携带该修饰,而且H2O2处理会导致上述tRNA的i6A37修饰水平降低。理论上,IMCRT tRNA-seq可以用于检测任何物种的tRNA i6A37修饰,为研究其生物学功能奠定坚实的基础。
我校生科院周志鹏教授、华中科技大学王锐教授和纽约州立大学Jia Sheng教授为论文共同通讯作者,我校学生陈莎莎(硕士)、李益楠(博士)和华中科技大学李源源(硕士,已毕业)、周红玲(博士)为共同第一作者。该研究中高通量测序及生物信息学分析由华中农业大学师生共同完成,化学合成等部分由华中科技大学师生完成。该研究成果得到了国家自然科学基金面上项目、华中农业大学新进高层次人才科研启动基金和中央高校基本科研业务费等项目的资助。
【英文摘要】
Chemical modifications in RNAs play crucial roles in diversifying their structures and regulating numerous biochemical processes. Since the 1990s, several hydrophobic prenyl-modifications have been discovered in various RNAs. Prenyl groups serve as precursors for terpenes and many other biological molecules. The processes of prenylation in different macromolecules have been extensively studied. We introduce here a novel chemical biology toolkit that not only labels i6A, a prenyl-modified RNA residue, by leveraging the unique reactivity of the prenyl group, but also provides a general strategy to incorporate fluorescence functionalities into RNAs for molecular tracking purposes. Our findings revealed that iodine-mediated cyclization reactions of the prenyl group occur rapidly, transforming i6A from a hydrogen-bond acceptor to a donor. Based on this reactivity, we developed an Iodine-Mediated Cyclization and Reverse Transcription (IMCRT) tRNA-seq method, which can profile all nine endogenous tRNAs containing i6A residues in Saccharomyces cerevisiae with single-base resolution. Furthermore, under stress conditions, we observed a decline in i6A levels in budding yeast, accompanied by significant decrease of mutation rate at A37 position. Thus, the IMCRT tRNA-seq method not only permits semi-quantification of i6A levels in tRNAs but also holds potential for transcriptome-wide detection and analysis of various RNA species containing i6A modifications.
